1 Introduction
This is a simple tutorial for electrodes localization. For more information and examples, please refer to Fieldtrip tutorials. Matlab, SPM, and Fieldtrip are required for this procedure. After adding the directory of Fieldtrip toolbox to searching path, you need to initialize it by running the command ft_defaults as well. It is convenient to add this command to startup.m, so that it will be executed automatically when starting Matlab.
2 Overview
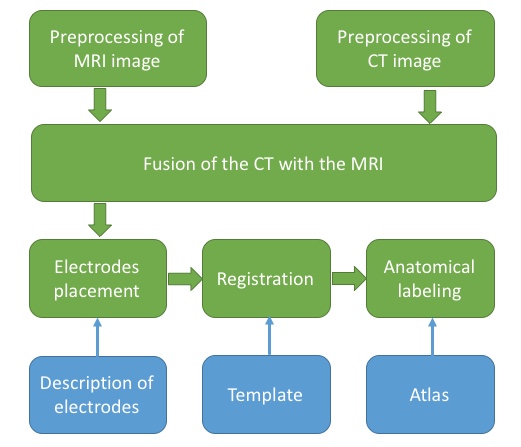
3 Preprocessing of MRI image
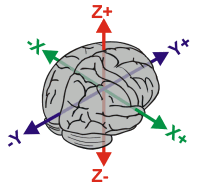
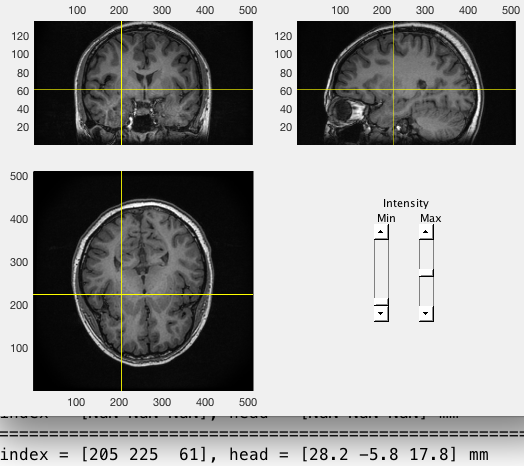
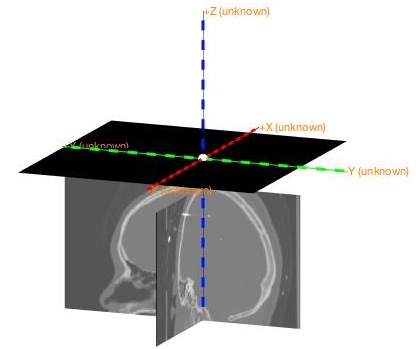
Load MRI and check its coordinate system first. As shown below,
- X should be the left/right dimension,
- Y should be the posterior/anterior dimension,
- Z should be the ventral/dorsal dimension.
% load MRI image
subjID = 's01';
filepath='data_folder';
mri = ft_read_mri([filepath '/' subjID '_MRI.nii']);
% determine left and right
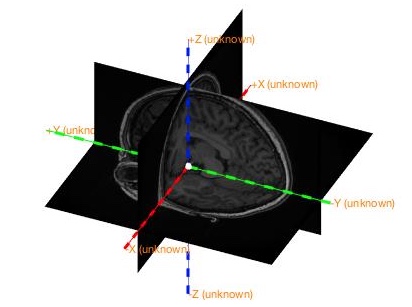
ft_determine_coordsys(mri);Check if there is a mismatch between the image and the labels of axis. Enter “n” in the command window of Matlab if it is correct. Otherwise, enter “y” and then following the instructions displayed in the command window.
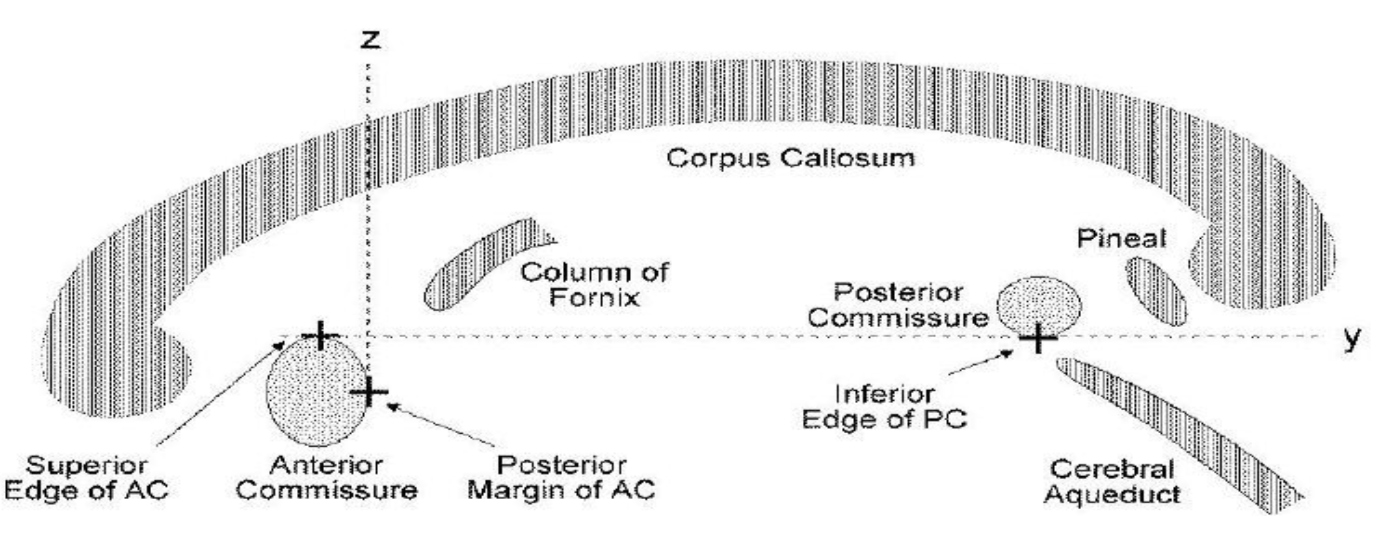
Then launch the interactive program to realign the image to ac-pc system.
cfg = [];
cfg.method = 'interactive';
cfg.coordsys = 'acpc';
mri_acpc = ft_volumerealign(cfg, mri);You need to identify four landmarks and press corresponding keys to assign coordinates to them. For more details, you can watch this video.
- key A: Anterior commissure (AC)
- key P: Posterior commissure (PC)
- key Z: Interhemispheric point along the positive midline
- key R: A point in the right hemisphere
Note that sometimes the display of the image is flipped here, so pay attention to the coordinates displayed in the command window to determine the left and right side of the image.
Save the preprocessed anatomical MRI to a nii file.
cfg = [];
cfg.filename = [filepath '/' subjID '_MRI_acpc'];
cfg.filetype = 'nifti';
cfg.parameter = 'anatomy';
ft_volumewrite(cfg, mri_acpc);4 Preprocessing of CT image
Similarly, CT image should also be realigned to ac-pc, which can also be achieved by select several landmarks (see Fieldtrip tutorials). However, selection may be inaccurate if the CT image is oblique, which will affect the fusion of CT and MRI images. Therefore, it is better to reorient the CT image in SPM.
Open SPM and click Display, then choose the CT image and click Done to open it. Note that the original image will be overwrite when saving the reoriented image, so it is better to make a copy of the original image.
% open spm
spm fmri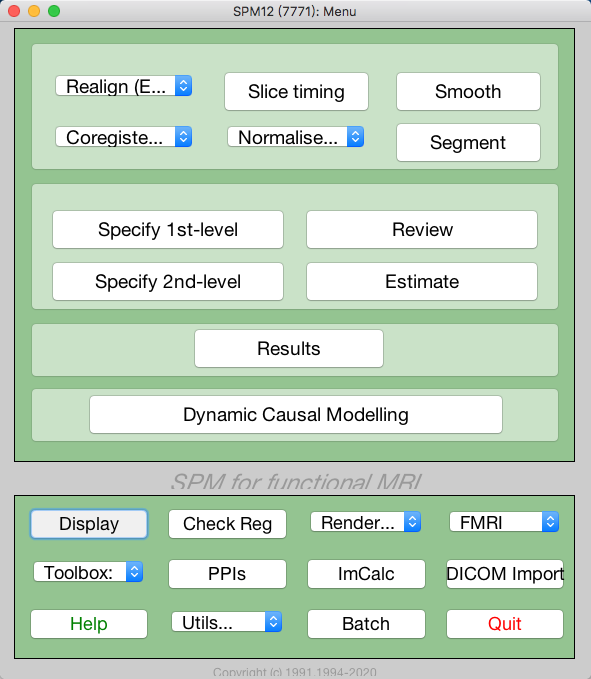
Reorient the image to match the orientation of MRI image by entering values for pitch, row, and yaw, and then set the origin to AC (approximately).
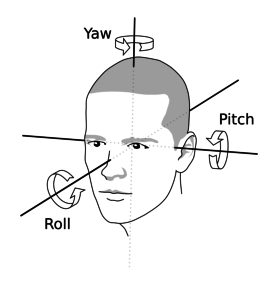
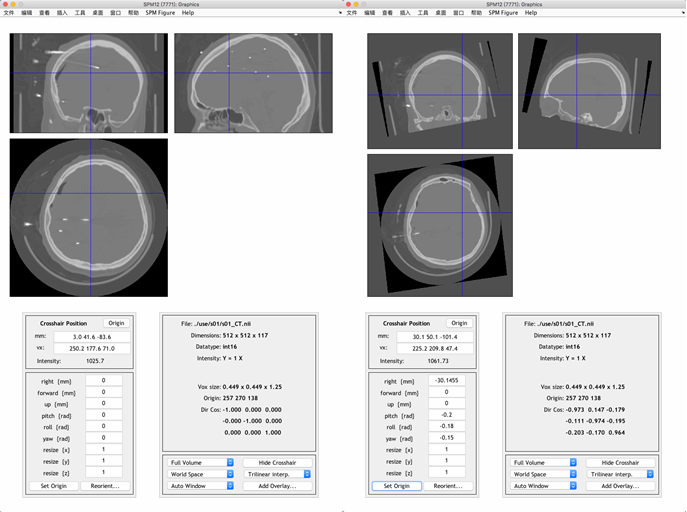 To save the reoriented image, click Reorient and then click Done.
To save the reoriented image, click Reorient and then click Done.
5 Fusion of the CT with the MRI
Align the reoriented CT image ct_acpc with acpc-aligned MRI image fsmri_acpc by the function ft_volumerealign.
% load spm reoriented img
ct_acpc = ft_read_mri([filepath '/' subjID '_CT_acpc.nii']);
% load acpc-aligned MRI image
fsmri_acpc = ft_read_mri([filepath '/' subjID '_MRI_acpc.nii']);
fsmri_acpc.coordsys = 'acpc';
% fusion
cfg = [];
cfg.method = 'spm';
cfg.spmversion = 'spm12';
cfg.coordsys = 'acpc';
cfg.viewresult = 'yes';
ct_acpc_f = ft_volumerealign(cfg, ct_acpc, fsmri_acpc);Check the results carefully to ensure that the CT image (shown in blue) matches with the MRI image (shown in red).
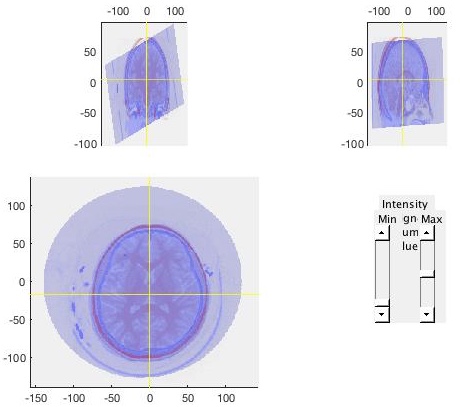
6 Electrodes placement
The electrode labels can be loaded from the header or generated by yourself, which will appear as a to-do list on the interactive electrode placement program.
% load labels from header
% hdr = ft_read_header([filepath filesep filename '.edf']);
% generate labels
ename=['F' 'G' 'Q' 'R'];
enum=[14 12 12 14];
names=cell(sum(enum),1);
for i=1:length(ename)
name_tmp=reshape(sprintf([ename(i) '%02d'],1:enum(i)),3,[])';
names(sum(enum(1:i))-enum(i)+1:sum(enum(1:i)),1)=cellstr(name_tmp);
end
hdr.label=names;Then, launch the program by providing electrode labels cfg.channel, CT image ct_acpc_f and MRI image fsmri_acpc to ft_electrodeplacement.
% place electrodes
cfg = [];
cfg.channel = hdr.label;
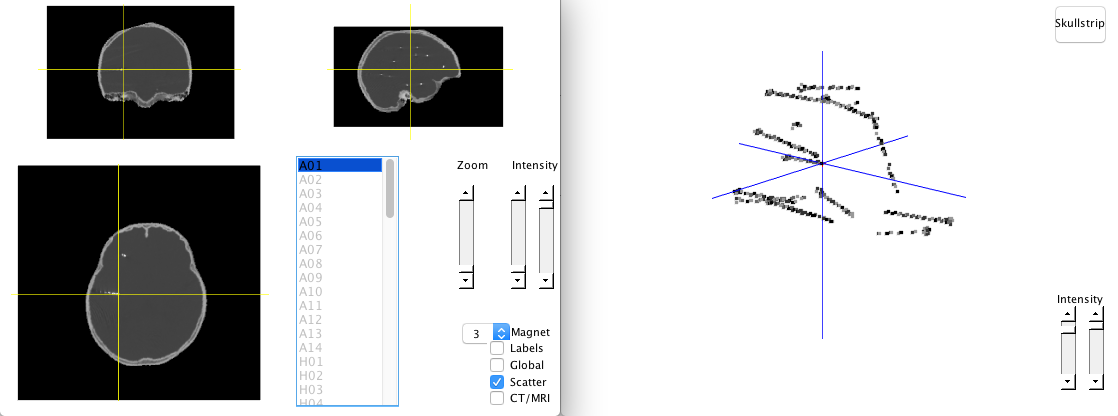
elec_acpc_f = ft_electrodeplacement(cfg, ct_acpc_f, fsmri_acpc);To assign a coordinate to an electrode, you need to select its label in the list first, and the click the corresponding electrode in the image, which appears to be light dots with fix interval in CT image. After assigning a coordinate, the label will change from gray to black, and a red cross will be shown on the image. To cancel a assignment, you should double click on the electrode label in the list. There are some options that can help you identify the electrodes.
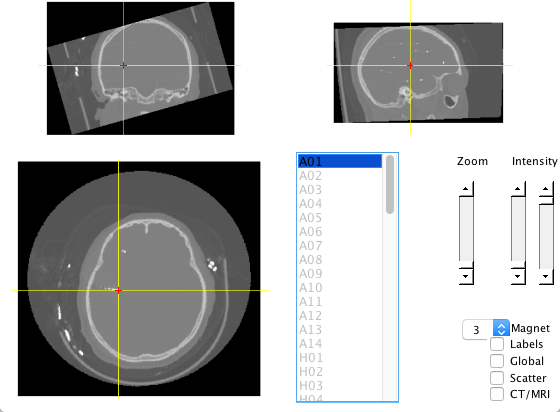
- Zoom: Zoom in the image to see the electodes.
- Intensity: Controls the contrast of the image.
- Magnet: Detect the weighted peak (
cfg.magtype = peakweighted) around 3 (cfg.magradius = 3) pixels by default. - Labels: Show labels on the image.
- Global: Display red crosses and labels wherever you are.
- CT/MRI: Switch to MRI image.
- Scatter: Plot the scatter image.
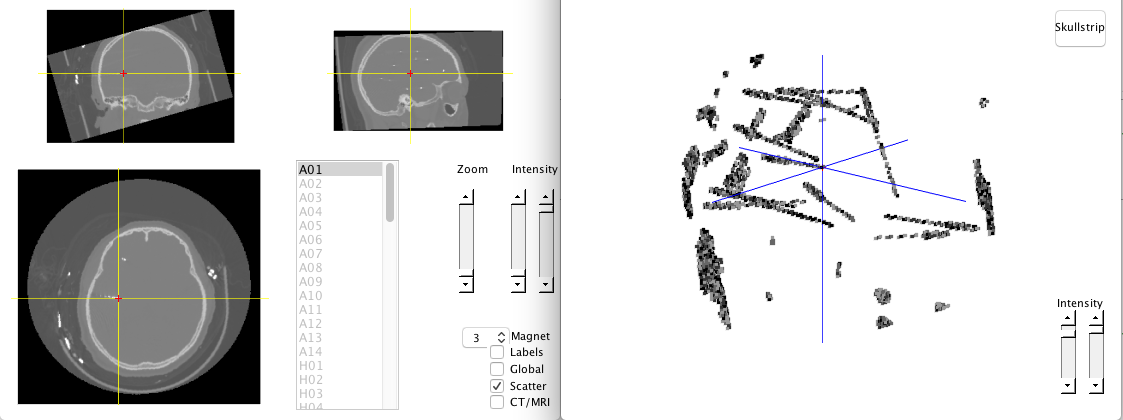
There is a skullstrip button in scatter mode, which can remove the skull so that electrodes can be easily identified. Electrodes can be select by data cursor in scatter mode as well.
You can visualize the electrodes and check if they are appropriate. If any error occurs, you can relaunch the placing program by providing previous results elec_acpc_f.
% visualize electrodes
ft_plot_ortho(fsmri_acpc.anatomy, 'transform', fsmri_acpc.transform, 'style', 'intersect');
ft_plot_sens(elec_acpc_f, 'label', 'on', 'fontcolor', 'w');
% place again if find any error
cfg = [];
cfg.elec = elec_acpc_f;
elec_acpc_f = ft_electrodeplacement(cfg, ct_acpc_f, fsmri_acpc);7 Volume-based registration
To make the electrode coordinates comparable across subjects, the individual brain need to be registered to the standard MNI brain. The volume-based registration considers the overall geometry of the individual brain and the template, so you may encounter a mismatch when the template is not suitable. For example, the default template is derived from the brains of adults, which may not suitable for children. Pediatric atlases provides a series of age-specific MRI brain templates for pediatric neuroimaging.
cfg = [];
cfg.nonlinear = 'yes';
cfg.spmversion = 'spm12';
cfg.spmmethod = 'new';
% you can specify a template by cfg.template
% cfg.template = 'path_to_the_template';
fsmri_mni = ft_volumenormalise(cfg, fsmri_acpc);The normalized image fsmri_mni can be saved in nifti format. To obtain electrode positions in standard MNI space, use ft_warp_apply to transform positions with deformation parameters contained in fsmri_mni.
% save normalized image
cfg = [];
cfg.filename = [filepath '/' subjID '_MRI_mni'];
cfg.filetype = 'nifti';
cfg.parameter = 'anatomy';
ft_volumewrite(cfg, fsmri_mni);
% obtain electrode positions in standard MNI space
elec_mni_fv = elec_acpc_f;
elec_mni_fv.elecpos = ft_warp_apply(fsmri_mni.params, elec_acpc_f.elecpos, 'individual2sn');
elec_mni_fv.chanpos = ft_warp_apply(fsmri_mni.params, elec_acpc_f.chanpos, 'individual2sn');
elec_mni_fv.coordsys = 'mni';
% Save normalized electrodes
save([subjID '_elec_mni_fv.mat'], 'elec_mni_fv');8 Anatomical labeling
Fieldtrip provides a function ft_volumelookup which can look up coordinates in an atlas loaded by ft_read_atlas. AAl and other atlases can be found in the “atlas” directory located in the path of Fieldtip. Brainnetome atlas seems more accurate than AAL atlas. The result returned by ft_volumelookup is a structure array, indicating the index of atlas labels for each coordinate. Therefore, you need to use these indexes to find the label names.
% load coordinates
load([filepath '/' subjID '_elec_mni_fv.mat']);
[ftver, ftpath] = ft_version;
% use brainnetome atlas
tname = 'template/atlas/brainnetome/BNA_MPM_thr25_1.25mm.nii';
atlas = ft_read_atlas([ftpath filesep tname]);
cfg = [];
cfg.roi = elec_mni_fv.chanpos(:,:);
cfg.atlas = atlas;
cfg.output = 'multiple';
labels = ft_volumelookup(cfg, atlas);
% save labels to the cell "position"
len=length(labels);
position=cell(len,1);
for i=1:len
[~, indx] = max(labels(i).count);
position{i}=labels(i).name(indx);
end